Info
openproblems_
Luecken et al. (2021)
7.78 GiB
14-02-2024
69249 cells × 13431 genes
Quick links
Used in
No related benchmarks found.
Single-cell Multiome (GEX+ATAC) data collected from bone marrow mononuclear cells of 12 healthy human donors.
CREATED
14-02-2024
DIMENSIONS
69249 × 13431
Single-cell CITE-Seq data collected from bone marrow mononuclear cells of 12 healthy human donors using the 10X Multiome Gene Expression and Chromatin Accessibility kit. The dataset was generated to support Multimodal Single-Cell Data Integration Challenge at NeurIPS 2021. Samples were prepared using a standard protocol at four sites. The resulting data was then annotated to identify cell types and remove doublets. The dataset was designed with a nested batch layout such that some donor samples were measured at multiple sites with some donors measured at a single site.
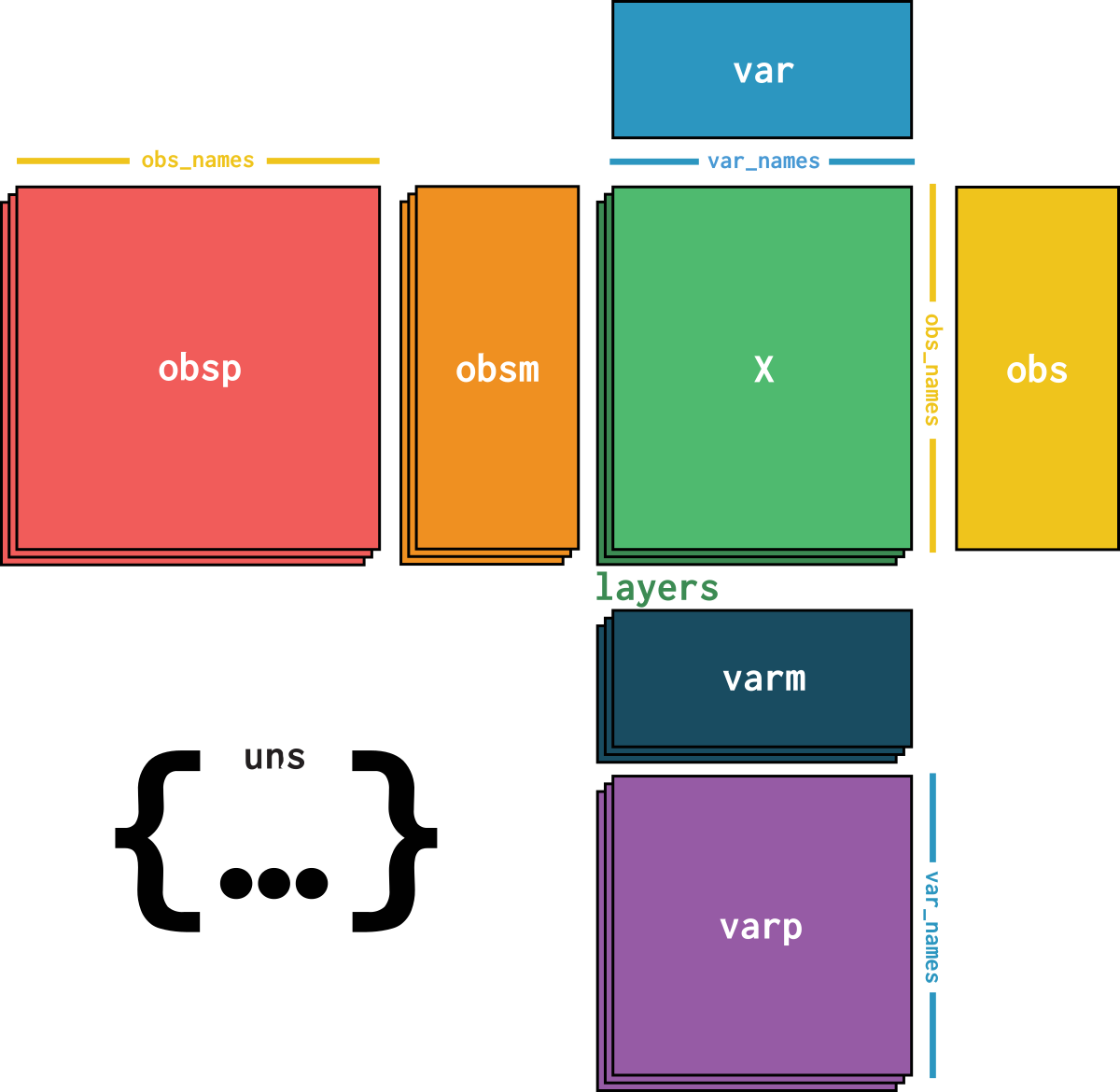
dataset_mod1 is an AnnData object with n_obs × n_vars = 69249 × 13431 with slots:
size_factors, cell_type, batchfeature_name, feature_id, hvg, hvg_scoreX_svdcounts, normalizeddataset_description, dataset_id, dataset_name, dataset_organism, dataset_reference, dataset_summary, dataset_url, normalization_iddataset_mod2 is an AnnData object with n_obs × n_vars = 69249 × 116490 with slots:
cell_type, batch, size_factorsfeature_name, feature_id, hvg, hvg_scoreX_svdcounts, normalizeddataset_description, dataset_id, dataset_name, dataset_organism, dataset_reference, dataset_summary, dataset_url, normalization_id| Name | Description | Type | Data type | Size |
|---|---|---|---|---|
| obs | ||||
batch
|
A batch identifier. This label is very context-dependent and may be a combination of the tissue, assay, donor, etc. |
vector
|
category
|
69249 |
cell_
|
Classification of the cell type based on its characteristics and function within the tissue or organism. |
vector
|
category
|
69249 |
size_
|
The size factors created by the normalisation method, if any. |
vector
|
float32
|
69249 |
| var | ||||
feature_
|
Unique identifier for the feature, usually a ENSEMBL gene id. |
vector
|
object
|
13431 |
feature_
|
A human-readable name for the feature, usually a gene symbol. |
vector
|
object
|
13431 |
hvg
|
Whether or not the feature is considered to be a ‘highly variable gene’ |
vector
|
bool
|
13431 |
hvg_
|
A ranking of the features by hvg. |
vector
|
float64
|
13431 |
| obsm | ||||
X_
|
The resulting SVD embedding. |
densematrix
|
float32
|
69249 × 100 |
| layers | ||||
counts
|
Raw counts |
sparsematrix
|
float32
|
69249 × 13431 |
normalized
|
Normalised expression values |
sparsematrix
|
float32
|
69249 × 13431 |
| uns | ||||
dataset_
|
Long description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
|
atomic
|
str
|
1 |
dataset_
|
A human-readable name for the dataset. |
atomic
|
str
|
1 |
dataset_
|
The organism of the sample in the dataset. |
atomic
|
str
|
1 |
dataset_
|
Bibtex reference of the paper in which the dataset was published. |
atomic
|
str
|
1 |
dataset_
|
Short description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
Link to the original source of the dataset. |
atomic
|
str
|
1 |
normalization_
|
Which normalization was used |
atomic
|
str
|
1 |
| Name | Description | Type | Data type | Size |
|---|---|---|---|---|
| obs | ||||
batch
|
A batch identifier. This label is very context-dependent and may be a combination of the tissue, assay, donor, etc. |
vector
|
category
|
69249 |
cell_
|
Classification of the cell type based on its characteristics and function within the tissue or organism. |
vector
|
category
|
69249 |
size_
|
The size factors created by the normalisation method, if any. |
vector
|
float32
|
69249 |
| var | ||||
feature_
|
Unique identifier for the feature, usually a ENSEMBL gene id. |
vector
|
object
|
116490 |
feature_
|
A human-readable name for the feature, usually a gene symbol. |
vector
|
object
|
116490 |
hvg
|
Whether or not the feature is considered to be a ‘highly variable gene’ |
vector
|
bool
|
116490 |
hvg_
|
A ranking of the features by hvg. |
vector
|
float64
|
116490 |
| obsm | ||||
X_
|
The resulting SVD embedding. |
densematrix
|
float32
|
69249 × 100 |
| layers | ||||
counts
|
Raw counts |
sparsematrix
|
float32
|
69249 × 116490 |
normalized
|
Normalised expression values |
sparsematrix
|
float32
|
69249 × 116490 |
| uns | ||||
dataset_
|
Long description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
|
atomic
|
str
|
1 |
dataset_
|
A human-readable name for the dataset. |
atomic
|
str
|
1 |
dataset_
|
The organism of the sample in the dataset. |
atomic
|
str
|
1 |
dataset_
|
Bibtex reference of the paper in which the dataset was published. |
atomic
|
str
|
1 |
dataset_
|
Short description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
Link to the original source of the dataset. |
atomic
|
str
|
1 |
normalization_
|
Which normalization was used |
atomic
|
str
|
1 |
dataset_mod1.layers['counts']In R: dataset_mod1$layers[["counts"]]
Type: sparsematrix, data type: float32, shape: 69249 × 13431
Raw counts
dataset_mod1.layers['normalized']In R: dataset_mod1$layers[["normalized"]]
Type: sparsematrix, data type: float32, shape: 69249 × 13431
Normalised expression values
dataset_mod1.obs['size_factors']In R: dataset_mod1$obs[["size_factors"]]
Type: vector, data type: float32, shape: 69249
The size factors created by the normalisation method, if any.
dataset_mod1.obs['cell_type']In R: dataset_mod1$obs[["cell_type"]]
Type: vector, data type: category, shape: 69249
Classification of the cell type based on its characteristics and function within the tissue or organism.
dataset_mod1.obs['batch']In R: dataset_mod1$obs[["batch"]]
Type: vector, data type: category, shape: 69249
A batch identifier. This label is very context-dependent and may be a combination of the tissue, assay, donor, etc.
dataset_mod1.obsm['X_svd']In R: dataset_mod1$obsm[["X_svd"]]
Type: densematrix, data type: float32, shape: 69249 × 100
The resulting SVD embedding.
dataset_mod1.uns['dataset_description']In R: dataset_mod1$uns[["dataset_description"]]
Type: atomic, data type: str, shape: 1
Long description of the dataset.
dataset_mod1.uns['dataset_id']In R: dataset_mod1$uns[["dataset_id"]]
Type: atomic, data type: str, shape: 1
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
dataset_mod1.uns['dataset_name']In R: dataset_mod1$uns[["dataset_name"]]
Type: atomic, data type: str, shape: 1
A human-readable name for the dataset.
dataset_mod1.uns['dataset_organism']In R: dataset_mod1$uns[["dataset_organism"]]
Type: atomic, data type: str, shape: 1
The organism of the sample in the dataset.
dataset_mod1.uns['dataset_reference']In R: dataset_mod1$uns[["dataset_reference"]]
Type: atomic, data type: str, shape: 1
Bibtex reference of the paper in which the dataset was published.
dataset_mod1.uns['dataset_summary']In R: dataset_mod1$uns[["dataset_summary"]]
Type: atomic, data type: str, shape: 1
Short description of the dataset.
dataset_mod1.uns['dataset_url']In R: dataset_mod1$uns[["dataset_url"]]
Type: atomic, data type: str, shape: 1
Link to the original source of the dataset.
dataset_mod1.uns['normalization_id']In R: dataset_mod1$uns[["normalization_id"]]
Type: atomic, data type: str, shape: 1
Which normalization was used
dataset_mod1.var['feature_name']In R: dataset_mod1$var[["feature_name"]]
Type: vector, data type: object, shape: 13431
A human-readable name for the feature, usually a gene symbol.
dataset_mod1.var['feature_id']In R: dataset_mod1$var[["feature_id"]]
Type: vector, data type: object, shape: 13431
Unique identifier for the feature, usually a ENSEMBL gene id.
dataset_mod1.var['hvg']In R: dataset_mod1$var[["hvg"]]
Type: vector, data type: bool, shape: 13431
Whether or not the feature is considered to be a ‘highly variable gene’
dataset_mod1.var['hvg_score']In R: dataset_mod1$var[["hvg_score"]]
Type: vector, data type: float64, shape: 13431
A ranking of the features by hvg.
dataset_mod2.layers['counts']In R: dataset_mod2$layers[["counts"]]
Type: sparsematrix, data type: float32, shape: 69249 × 116490
Raw counts
dataset_mod2.layers['normalized']In R: dataset_mod2$layers[["normalized"]]
Type: sparsematrix, data type: float32, shape: 69249 × 116490
Normalised expression values
dataset_mod2.obs['cell_type']In R: dataset_mod2$obs[["cell_type"]]
Type: vector, data type: category, shape: 69249
Classification of the cell type based on its characteristics and function within the tissue or organism.
dataset_mod2.obs['batch']In R: dataset_mod2$obs[["batch"]]
Type: vector, data type: category, shape: 69249
A batch identifier. This label is very context-dependent and may be a combination of the tissue, assay, donor, etc.
dataset_mod2.obs['size_factors']In R: dataset_mod2$obs[["size_factors"]]
Type: vector, data type: float32, shape: 69249
The size factors created by the normalisation method, if any.
dataset_mod2.obsm['X_svd']In R: dataset_mod2$obsm[["X_svd"]]
Type: densematrix, data type: float32, shape: 69249 × 100
The resulting SVD embedding.
dataset_mod2.uns['dataset_description']In R: dataset_mod2$uns[["dataset_description"]]
Type: atomic, data type: str, shape: 1
Long description of the dataset.
dataset_mod2.uns['dataset_id']In R: dataset_mod2$uns[["dataset_id"]]
Type: atomic, data type: str, shape: 1
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
dataset_mod2.uns['dataset_name']In R: dataset_mod2$uns[["dataset_name"]]
Type: atomic, data type: str, shape: 1
A human-readable name for the dataset.
dataset_mod2.uns['dataset_organism']In R: dataset_mod2$uns[["dataset_organism"]]
Type: atomic, data type: str, shape: 1
The organism of the sample in the dataset.
dataset_mod2.uns['dataset_reference']In R: dataset_mod2$uns[["dataset_reference"]]
Type: atomic, data type: str, shape: 1
Bibtex reference of the paper in which the dataset was published.
dataset_mod2.uns['dataset_summary']In R: dataset_mod2$uns[["dataset_summary"]]
Type: atomic, data type: str, shape: 1
Short description of the dataset.
dataset_mod2.uns['dataset_url']In R: dataset_mod2$uns[["dataset_url"]]
Type: atomic, data type: str, shape: 1
Link to the original source of the dataset.
dataset_mod2.uns['normalization_id']In R: dataset_mod2$uns[["normalization_id"]]
Type: atomic, data type: str, shape: 1
Which normalization was used
dataset_mod2.var['feature_name']In R: dataset_mod2$var[["feature_name"]]
Type: vector, data type: object, shape: 116490
A human-readable name for the feature, usually a gene symbol.
dataset_mod2.var['feature_id']In R: dataset_mod2$var[["feature_id"]]
Type: vector, data type: object, shape: 116490
Unique identifier for the feature, usually a ENSEMBL gene id.
dataset_mod2.var['hvg']In R: dataset_mod2$var[["hvg"]]
Type: vector, data type: bool, shape: 116490
Whether or not the feature is considered to be a ‘highly variable gene’
dataset_mod2.var['hvg_score']In R: dataset_mod2$var[["hvg_score"]]
Type: vector, data type: float64, shape: 116490
A ranking of the features by hvg.