Info
zenodo_
Stickels et al. (2020)
98.93 MiB
23-09-2024
29309 cells × 8442 genes
Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2
zenodo_
Stickels et al. (2020)
98.93 MiB
23-09-2024
29309 cells × 8442 genes
DATASET ID
zenodo_spatial/slideseqv2/mouse_hippocampus_puck
REFERENCE
Stickels et al. (2020)
SIZE
98.93 MiB
CREATED
23-09-2024
DIMENSIONS
29309 × 8442
Gene expression library of mouse hippocampus puck profiled using Slide-seq V2.
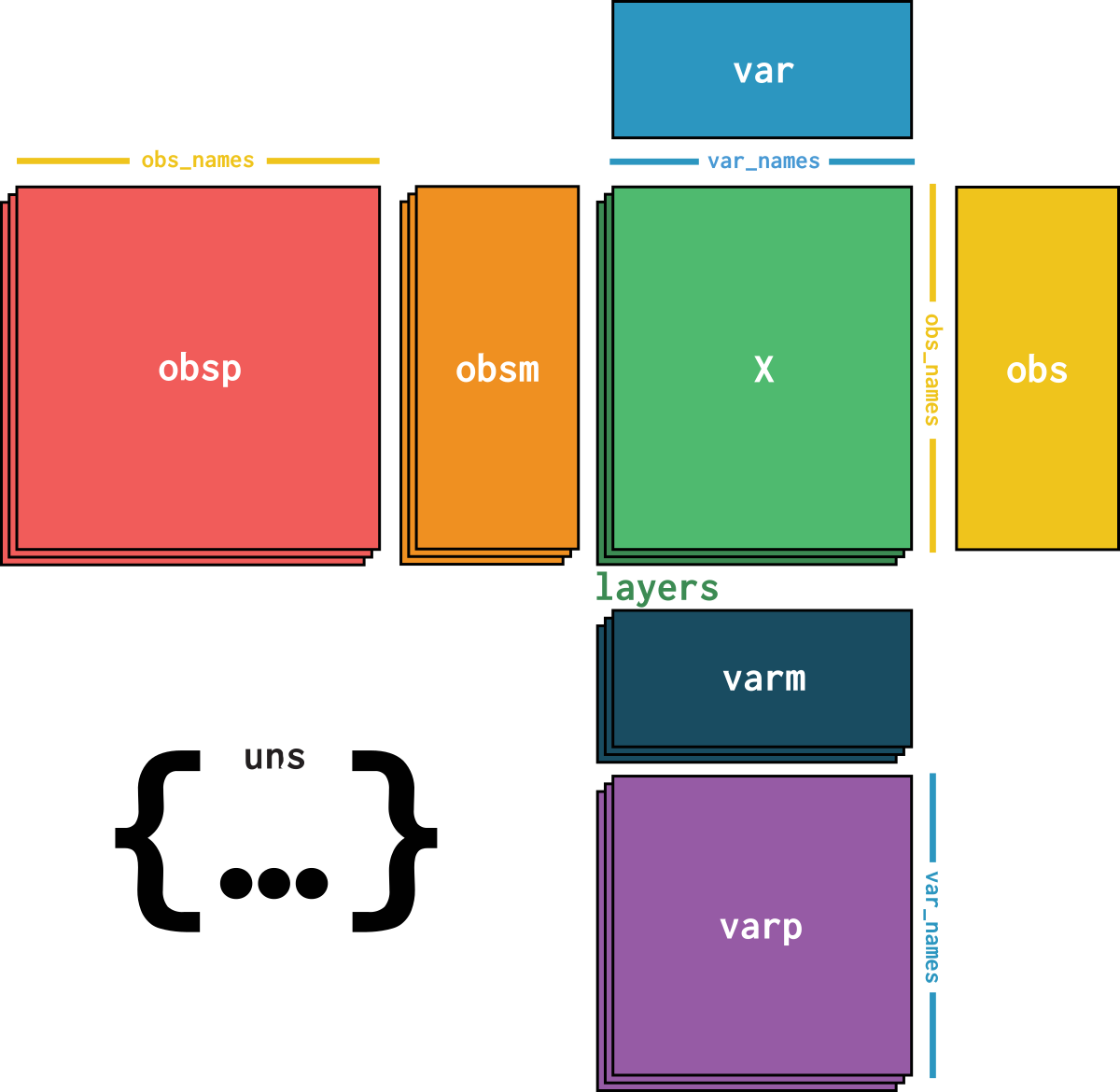
dataset is an AnnData object with n_obs × n_vars = 29309 × 8442 with slots:
feature_namecountsdataset_description, dataset_id, dataset_name, dataset_organism, dataset_reference, dataset_summary, dataset_url| Name | Description | Type | Data type | Size |
|---|---|---|---|---|
| var | ||||
feature_
|
A human-readable name for the feature, usually a gene symbol. |
vector
|
object
|
8442 |
| layers | ||||
counts
|
Raw counts |
sparsematrix
|
int64
|
29309 × 8442 |
| uns | ||||
dataset_
|
Long description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
|
atomic
|
str
|
1 |
dataset_
|
A human-readable name for the dataset. |
atomic
|
str
|
1 |
dataset_
|
The organism of the sample in the dataset. |
atomic
|
str
|
1 |
dataset_
|
Bibtex reference of the paper in which the dataset was published. |
atomic
|
str
|
1 |
dataset_
|
Short description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
Link to the original source of the dataset. |
atomic
|
str
|
1 |
dataset.layers['counts']In R: dataset$layers[["counts"]]
Type: sparsematrix, data type: int64, shape: 29309 × 8442
Raw counts
dataset.uns['dataset_description']In R: dataset$uns[["dataset_description"]]
Type: atomic, data type: str, shape: 1
Long description of the dataset.
dataset.uns['dataset_id']In R: dataset$uns[["dataset_id"]]
Type: atomic, data type: str, shape: 1
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
dataset.uns['dataset_name']In R: dataset$uns[["dataset_name"]]
Type: atomic, data type: str, shape: 1
A human-readable name for the dataset.
dataset.uns['dataset_organism']In R: dataset$uns[["dataset_organism"]]
Type: atomic, data type: str, shape: 1
The organism of the sample in the dataset.
dataset.uns['dataset_reference']In R: dataset$uns[["dataset_reference"]]
Type: atomic, data type: str, shape: 1
Bibtex reference of the paper in which the dataset was published.
dataset.uns['dataset_summary']In R: dataset$uns[["dataset_summary"]]
Type: atomic, data type: str, shape: 1
Short description of the dataset.
dataset.uns['dataset_url']In R: dataset$uns[["dataset_url"]]
Type: atomic, data type: str, shape: 1
Link to the original source of the dataset.
dataset.var['feature_name']In R: dataset$var[["feature_name"]]
Type: vector, data type: object, shape: 8442
A human-readable name for the feature, usually a gene symbol.